This is an old revision of the document!
Selection of linkers for the DNA-bindig ligand
Objective:
Find the aliphatic linker for the two pyrrole-carboxamide ligands bound in the narrow groove of DNA.
Need to get a dual ligand binds to a 5'-gaagcttc-3' DNA sequence.
The work done on the Abalone program.
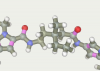
Pyrrolecarboxamide ligand
The original ligand was drawn in the graphics window with the mouse. Then, the button Make ad hoc model has been pressed that led to the creation of the following model:
The procedure took 7 minutes1). This is a very crude model. All valence parameters taken from the universal force field. Electrostatics was calculated very roughly (you can expect a 10-20% error in the dipole moment). Nevertheless, it is quite adequate to the task. Moreover, this model is much better than we could obtain using a semi-empirical methods and/or Mulliken charges.
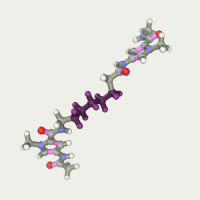
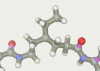
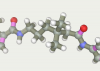
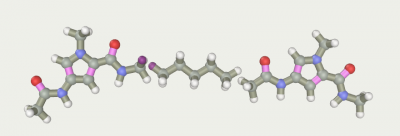
Aliphatic linker was made in the “Build chain” editor. We took him long enough in order to try different truncated versions by isomerizing it. Adding two pyrrolecarboxamide fragments we have the desired ligand:
 Fragments placed against each other
Fragments placed against each other
and the hydrogen which will be a connection was selected.
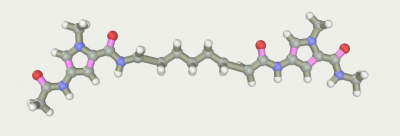 Gether the fragments2).
Gether the fragments2).
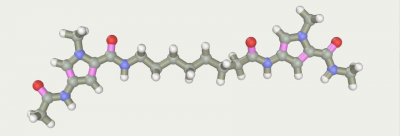 A short optimization3) to eliminate tension.
A short optimization3) to eliminate tension.
Complex
The model of DNA has been collected in the “Build сhain” editor. In order to avoid large deformations on the first phase we worked with the “frozen” model.
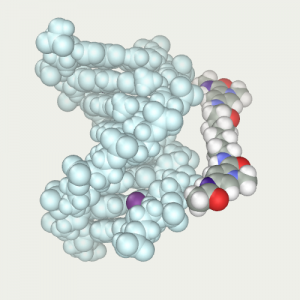 Ligand is positioned in front of DNA and restrained to tighten it in the narrow groove | 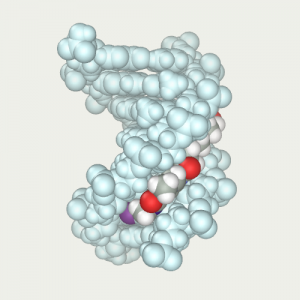 Ligand was tightened by means of dynamics at low temperature 4) | 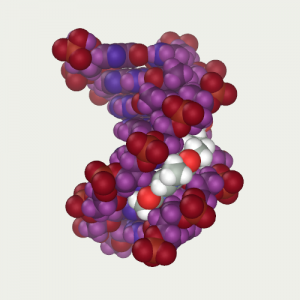 Thawed DNA and repeated the protocol optimization by dynamics (B).js |
Calculations were carried out in the AMBER99 force field in implicit water (Sheffield method).
Selection of the linker
In order to determine the most suitable linker we are going over its isomers. To do this, we have selected a place for isomerization and used the batch processing/find linker.js protocol. This protocol is similar to optimization by dynamics (B).js but it performs multiple optimization after isomerization of selected part of the molecule.
During the night the program scanned the 200 variants of linkers. Consider the top 10, according to the energy of interaction between DNA and the ligand.
| Attempt | Intermolecular Energy, kcal/mol | |
|---|---|---|
| 60 | -119.53 | 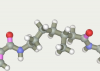 |
| 9 | -119.16 | 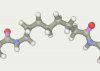 |
| 59 | -118.59 | 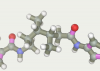 |
| 163 | -118.32 | 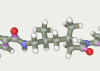 |
| 79 | -117.81 | 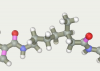 |
| 124 | -117.74 | 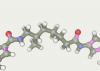 |
| 121 | -117.61 | 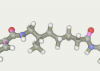 |
| 76 | -117.59 | 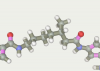 |
| 167 | -117.52 | 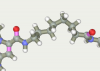 |
| 164 | -117.49 | 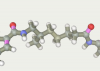 |
The top ten twice entered the original line-linker (attempts 9 и 167). Three times the present 60, 79 and 76 as well as 124, 121 and 164. Versions 59 and 163 present one time. So in top 10 there are only 5 linkers versions. All variants have a linear section from 5 to 7 carbon atoms. Therefore, the shorter linkers do not suit us.
Now look at the best variants for the full potential of the system. It should be borne in mind that they are not saved by this criterion in the protocol find linker.js. If we really want to explore this question, it is necessary to supplement the protocol. However, look at the variants with the lowest total potential in the range of 2 kcal/mol. Such linkers were only 5. And these were the different variants:
So, we chose 15 out of 200 candidates for a more thorough investigation.
This is a case study. In the real work an order of magnitude more options would be considered.